 |
 |
- Search
| Chronobiol Med > Volume 1(1); 2019 > Article |
|
Abstract
Objective
Dividing bipolar disorder (BD) into clinical subphenotypes is a possible approach to determine the genetic basis of BD. The goal of this study was to find genes associated with BD by performing a genome-wide association study (GWAS) based on the subphenotypes, i.e., sleeplessness symptom of bipolar mania (SBM) vs. non-sleeplessness symptom of bipolar mania (NSBM).
Methods
A total of 2,191 cases, 1,434 controls, and 703,012 single nucleotide polymorphisms (SNPs) in the merged samples from the Translational Genomics Institute and the Genetic Association Information Network were investigated. In individuals with bipolar I disorder, a GWAS of SBM vs. NSBM was performed.
Results
We identified 44 trait-associated SNPs with p<10-4 in a case-only analysis. The most statistically significant peak p-value was observed for rs10492908 (odds ratio=0.1832; p=9.27×10-7; Permuted p=1.00×10-6), which is located within the region of the genome encoding WW domain-containing oxidoreductase (WWOX) on chromosome 16q23. A total of eight genomic regions of interest (ROIs) on chromosomes 3, 5, 8, 9, 12, and 17 were also identified, and ROR2 and ANKFN1 in two ROIs may be related with SBM.
Bipolar disorder (BD) is a common mental disorder characterized by episodic mood symptoms, including episodes of mania or hypomania and depression, and its mood episodes are severe and lifelong [1,2]. The lifetime prevalence of BD is approximately 3.9–6.4% in the general population [3-5], and the lifetime prevalence of bipolar I disorder (BID) is between 1% and 2% [6,7]. Relapse and recurrence are very common and the lifetime risk of suicide is as high as 20% in BD patients, resulting in serious economic costs [8].
The representative defining character of BD is the development of alternating episodes of mania, hypomania, or depression [6]. All BD episode types typically show prominent sleep disturbance. Usually, the necessity for sleep is reduced during mania or hypomania episodes, and patients typically suffer from insomnia or hypersomnia during major depressive episodes [9]. Several studies have suggested that sleep deprivation results in significantly increased negative mood [10,11]. Sleep disturbances may contribute to mood-related features across psychiatric disorders [12]. A bidirectional relationship between sleep dysfunction and symptoms of psychiatric disorders has been hypothesized, such that a vicious cycle of disturbance in mood and symptoms interrupt nighttime sleep, and this sleep deprivation contributes to mood regulation difficulties and symptom worsening [12,13]. Several findings have suggested that circadian rhythm dysfunction is associated with the pathophysiology of BD [14-17]. Commonly, BD patients have circadian rhythm-related clinical manifestations, such as a cyclical/seasonal pattern of relapse, activity changes, periodicity of manic-depressive episodes, diurnal variation in mood, alteration of the secretion of hormones and other endogenous substances, as well as sleep disturbances, such as hypersomnia or insomnia during depression and sleeplessness during mania [18]. Additionally, the interpersonal, environmental, and pharmacological events of BD may cause sleep deprivation [13,19]. Sleep disturbances can be caused by circadian dysfunctions in BD, and inversely, sleep disturbances may have an influence on emotional regulation [20]. Our group previously suggested circadian rhythm hypotheses to explain mixed features, antidepressant treatment resistance, and manic switching in BD, and we emphasized that circadian dysregulation may cause recurrent unipolar depression and BD [21-25]. Among the disturbance of sleep-wake rhythm, sleeplessness—reduced need for sleep—is a distinctive symptom in the bipolar mania. In the diagnostic criteria of BD, ‘decreased need for sleep’ is one of the main symptoms of the mania or hypomania [26]. Thus, the sleeplessness is worthy of consideration as a major subphenotype of bipolar mania.
Even though BD is a highly heritable psychiatric disorder, the investigation of specific genetic variations has suggested limited findings [27-30]. Since, it has been suggested that dividing BD into subgroups according to clinical phenotypes is a possible approach for further meaningful genetic studies in BD. Lett et al. [31] reported that ZNF804A and CACNA1C are associated with a psychotic subphenotype of BD. Our group previously performed a genome-wide association (GWA) analysis of BD using subphenotypes, such as seasonal pattern mania [32] and hypersomnia symptom of bipolar depression [33].
In the present study, we performed a GWA analysis of the sleeplessness symptom of bipolar mania (SBM) vs. non-sleeplessness symptom of bipolar mania (NSBM) in BD subjects and controls of European ancestry, genotyped as part of the Genetic Association Information Network (GAIN) by the Bipolar Genome Study (BiGS).
A total of 1,556 unrelated BID subjects of European ancestry were selected from those collected by the National Institute of Mental Health Genetics Initiative for Bipolar Disorder in 5 waves at 11 sites across the United States. Recruitment for waves 1–2 consisted of extended multiplex families with a BID proband, waves 3–4 included families with a BID proband and at least one other sibling with BID or schizoaffective disorder, bipolar type, and wave 5 consisted of unrelated BID cases. All subjects provided written informed consent according to the local Institutional Review Board protocols and were interviewed using the Diagnostic Interview for Genetic Studies (DIGS) [34]. Information sources for the study included both family informants and medical records. These information sources were reviewed in addition to the interview by a panel of experienced clinicians to reach a final best-estimate diagnosis. The full version of the DIGS interview contains over 2,000 questions with detailed information regarding manic and depressive episodes, including sleep patterns, and is available for these subjects. Control subjects were selected from those confirmed based on a National Institute of Mental Health-supported contract mechanism between Dr. Pablo Gejman and Knowledge Networks, Inc [35]. All subjects donated a blood sample and completed a medical questionnaire. The controls were matched with respect to ethnicity and gender to the BD cases, and all control subjects who had a history of BD, psychosis, or recurrent major depression were excluded from our study.
Initial samples were genotyped at the Broad Institute as part of GAIN using Affymetrix SNP Array 6.0 [Affymetrix, Santa Clara, CA, USA; 1M single nucleotide polymorphism (SNPs)]. A total of 1,001 BD cases, 1,033 controls, and 724,067 SNPs were available for analysis following an extensive quality control process to eliminate individuals with >10% missing data, SNPs with poor allele clustering, duplicate data, significant deviation from Hardy–Weinberg equilibrium at p<10-6, and minor allele frequencies <0.05. The second sample was genotyped at the Translational Genomic Institute (TGEN) and a comparable quality control process was used, resulting in 1,190 BD cases, 401 controls, and 728,187 SNPs for analysis. A total of 2,191 cases, 1,434 controls, and 703,012 SNPs in the merged samples of TGEN and GAIN were investigated.
Phenotypes were based on the information from the Bipolar Disorder Phenome Database, which is a resource for genetic studies that compiles data across the DIGS 2, 3, and 4 to obtain a common set of variables for samples from each subject [32,36]. As a part of the DIGS interview, BID subjects were queried about sleep during the most severe manic episode. Subjects were asked the question, “Did you need less sleep than usual?’’ BID subjects who answered ‘Yes’ were assigned to the SBM group and subjects who answered ‘No’ were assigned to the NSBM group. Those who answered ‘Unknown’ to the question were categorized as missing because their symptoms may be not related to sleep patterns. After categorization, there were 302 BID subjects in the SBM group and 84 subjects in the NSBM group.
To investigate the genetic factors contributing to SBM, a caseonly analysis of SBM vs. NSBM was performed. This association analysis was performed using logistic regression with the PLINK software package [37] and covariate adjustment for age and sex. Because a sampling bias owing to the selection of a small subset of individuals may result in spurious findings, adaptive permutations were performed to assess the significance of the results using PLINK. After SNP imputation, the same logistic regression analysis was repeated using the imputed sample for the functional enrichment analysis. A genome-wide significance threshold of p< 5×10-8 was used after correction for multiple testing issues [38].
Common linkage disequilibrium (LD) blocks were identified using the Haploview software based on D’ and r2, with default parameters and HapMap CEU+TSI(R2) [39]. Furthermore, proxy SNPs based on regional recombination rates were identified using SNAP and 1000 Genomes Pilot 1 data.
The genotypes for missing markers in a dataset can be extrapolated on the basis of the correlation between genotypes in a reference dataset and an LD analysis. Similar levels of association should be observed for genotyped markers and imputed markers. The IMPUTE2 tool [40] and the CEU panel of HapMap 3 + 1000 Genomes Pilot haplotypes as a reference were used for imputation.
A functional enrichment analysis was performed on the gene sets harboring the identified SNPs (p<0.005) using DAVID software [41]. The gene sets were derived from the results of a logistic regression analysis performed within the imputed sample. An annotation process was performed using the RefSeq database (https://www.ncbi.nlm.nih.gov/refseq/). R software version 3.0 (R Foundation for Statistical Computing, Vienna, Austria) was used for annotation, and each SNP was matched to a gene located within 100,000 base pairs. Significant functions were suggested based on enrichment scores.
We performed a GWA analysis to identify SNPs associated with SBM. Figure 1 shows a Manhattan plot representing the chromosomal view of the log p-values for the SNPs. We identified 44 traitassociated SNPs with p<10-4 in the case-only analysis, and the lowest p-value was observed for rs10492908 [odds ratio (OR)=0.1832; p=9.27× 10-7; Permuted P=1.00×10-6]. This SNP was located within the region of the genome encoding WW domain-containing oxidoreductase (WWOX) on chromosome 16q23.
We identified a total of eight genomic regions of interest (ROIs) on chromosomes 3, 5, 8, 9, 12, and 17 that contained at least two SNPs with p<10-4 and adequate support for an association (i.e., p< 10-3) for additional SNPs within 100 kb. The 5 ROIs, except ROI3 and ROI4, were located in 9 genes. Among these genes, seven included at least 3 SNPs, CNTN4 in ROI1, NCEH1, ECT2 in ROI2, ROR2 in ROI5, ANKFN1 in ROI7, and LINC01483 in ROI8 (Supplementary Table 1 in the online-only Data Supplement).
LD for the ROIs was calculated from HapMap CEU+TSI(R2) using Haploview software after imputing the data using genotypes from the publicly available CEU HapMap 3 + 1000 Genomes Pilot haplotypes data set. As shown in Figure 2, all eight ROIs were located within or near blocks of high LD. These results suggest that these regions have functional significance related to SBM. For the first ROI, rs7646671, which showed a peak p-value of 2.7×10-5, was located within the CNTN4 gene on 3p26.3, and six additional SNPs were observed with p-values of <10-3 within the same region (Figure 2A). In the second ROI, rs11706689, which showed a peak p-value of 4.26×10-5, was located within the ECT2 gene on 3q26, and four additional SNPs were detected in this ROI (p<10-3) (Figure 2B). In ROI6, rs10785430 had the lowest p-value of 4×10-5, and was located within the ADAMTS20 gene on 12q12 and six other SNPs (p<10-3) were detected in the ROI (Figure 2F). Although only a few SNPs within the other 5 ROIs were located in genes, these regions showed significance of p<10-3 in the association study. These results suggest that functional alterations of genes in ROIs may contribute to the occurrence of SBM.
We performed a functional enrichment analysis for associated SNPs after imputing the data using genotypes from the publicly available CEU HapMap 3 + 1000 Genomes Pilot haplotypes data set. We identified gene ontology functions for 3,078 genes containing SNPs demonstrating associations with p<0.005. As shown in Table 1, significant functional enrichments were observed for cell development, cell adhesion, signal transduction, cell recognition, synaptic transmission, and cell motility-related pathways in the SBM vs. NSBM analysis. These findings suggest that the expression of SBM in BD may be related to altered functions in the pathways of the enriched genes identified in this study.
Various investigations have searched for genetic components contributing to BD susceptibility. Although several GWA studies (GWASs) have suggested possible BD susceptibility genes, there has been little consistency among the results of GWA studies. The Wellcome Trust Case-Control Consortium found an association of rs420259 at chromosome 16p12 (p=6.3×10-8) with BD [42]. Another group found that diacylglycerol kinase eta (DGKH) is associated with BD in a combined GWAS sample (p=1.5×10-8) [43]. MYO5B (p=1.66×10-7) and TSPAN8 (p=6.11×10-7) were suggested to be associated with BD [27]. The Psychiatric GWAS Consortium Bipolar Disorder Working Group confirmed genome-wide significant evidence for an association between BD and CACNA1C (p=1.52× 10-8), which encodes the α subunit of the L-type voltage-gated calcium channel, and ODZ4 (p=4.40×10-8), which encodes a member of a family of cell surface proteins, the teneurins, in a combined GWAS sample [44].
To better understand the genetic basis underlying BD, we performed a GWA study of the SBM subphenotype, which may represent a genetically distinct and homogenous feature. The most statistically significant peak representing a GWA was observed for rs10492908 (OR=0.1832; p=9.27×10-7; Permuted P=1.00×10-6), which is located within WWOX on chromosome 16q23. WWOX has been implicated in human tumorigenesis, especially as a putative tumor suppressor [45]. WWOX has one COOH-terminal short-chain alcohol dehydrogenase/reductase and two N-terminal WW domains (containing conserved tryptophan residues). Since these domains mediate protein–protein interactions, WWOX binds to various proteins. Although WWOX is well-known with regard to cancer, it is also related to nervous system development and Alzheimer’s disease (AD) [46]. Chen et al. [47] reported that WWOX was differentially expressed during various stages of brain development in mice, implicating a potential role for WWOX in promoting neuronal differentiation and maturation. Mallaret et al. [48] showed that WWOX is mutated in autosomal recessive cerebellar ataxia with epilepsy and mental retardation. WWOX is also suggested to have a role in the pathogenesis of AD via tau and glycogen synthase kinase 3β (GSK3β). Tau is a microtubule-associated protein that plays a role in in microtubule assembly and stability. GSK3β has an important role in tau hyperphosphorylation at its microtubule-binding domains, resulting in a disruption of microtubule stability. Wang et al. [49] reported that WWOX can bind directly to GSK3β via its short-chain alcohol dehydrogenase/reductase domain and promotes neuronal differentiation via the negative regulation of GSK3β. GSK3β is a key kinase in several important signaling pathways and is involved in both BD and AD [50].
Lithium is a mood stabilizing medication that is prescribed to control BD. Several studies have suggested that lithium regulates circadian rhythms, and this regulation may mediate the therapeutic effects of lithium [51-53]. A study of chronic lithium administration in rats suggested that lithium influences the synchronization of circadian rhythms by affecting melatonin levels along the retinal-hypothalamic-pineal pathway [54]. Lithium directly and reversibly inhibits GSK3β [55,56]. Treatment of cells with lithium inhibits the GSK3β-dependent phosphorylation of the microtubule-associated protein tau [57]. WWOX, the most statistically significant peak observed in the present GWA study, may be related to BD.
For further evaluation, each gene in the ROIs was reviewed with respect to previously detected associations with psychiatric disorders. We found that receptor tyrosine kinase-like orphan receptor 2 (ROR2) in ROI5 and ANKFN1 in ROI7 may be involved in the sleepless SBM. In mammals, the Ror-family receptor tyrosine kinases include two structurally related proteins, ROR1 and ROR2. ROR1-ROR2 complexes modulate synapse formation in hippocampal neurons [58]. ROR1 is reported to be associated with insomnia [59] and BD [60]. In the developing mouse brain, ROR2 is expressed predominantly in the limbic neocortex, hippocampal neuroepithelium, and caudate putamen, although apparent expression of ROR1 has not been observed in the brain [61]. Ankyrin-repeat and fibronectin type III domain-containing 1 (ANKFN1) in ROI7 is a possible candidate gene based on a GWAS of cannabis dependence [62]. Moreover, we searched for enriched pathways for genes in the ROIs. CNTN4 in ROI1 belonged to all pathways in annotation clusters 1, 2, and 4 and cell motion in annotation cluster 6. ROR2 in ROI5 belonged to all pathways in annotation cluster 2.
Owing to the limitations of genetic studies of psychiatric disorders, we used a subphenotype GWA study to identify candidate genes. The heterogeneous phenotype (BD) was divided into clinically homogeneous subphenotypes, which may be more homogenous at the genetic level as well. Most genetic research related to BD based on subphenotypes has focused on age at onset, psychosis, and episode frequency. Accordingly, we considered SBM as a subphenotype of BD. Although WWOX, which was the most highly significant peak in the GWA did not reach the 5×10-8 threshold for statistical significance in a GWA study and was not observed in the genomic ROIs, this SNP may be associated with BD. A limitation of this study is that candidate genes known to be related to BD from previous studies were not replicated in our results. In the future, genetic studies using more detailed subphenotypes in a large sample of BD are expected to verify the findings of the present study and may suggest more meaningful associations.
Supplementary Materials
The online-only Data Supplement is available with this article at https://doi.org/10.33069/cim.2018.0002.
Supplementary Table 1.
ROIs with enriched association in the sleeplessness symptom of bipolar mania vs. non-sleeplessness symptom of bipolar mania analysis
Acknowledgments
This study was supported by the Korea Health 21 R&D Project funded by the Ministry of Health & Welfare, Republic of Korea (HM14C2606).
This study data was obtained from the Bipolar Genome Study (BiGS). Especially, we thank to Dr. Kelsoe from UCSD for giving study data.
NOTES
Conflicts of Interest
Author Contributions
Conceptualization: Heon-Jeong Lee. Data curation: Heon-Jeong Lee, Chul-Hyun Cho. Formal analysis: Ji-Hye Choi, Hyun Goo Woo. Investigation: Chul-Hyun Cho, Ji-Hye Choi. Methodology: Ji-Hye Choi, Hyun Goo Woo. Software: Ji-Hye Choi, Hyun Goo Woo. Supervision: Heon-Jeong Lee. Validation: Heon-Jeong Lee, Hyun Goo Woo. Visualization: Chul-Hyun Cho, Ji-Hye Choi. Writing—original draft: Chul-Hyun Cho, Ji-Hye Choi. Writing—review & editing: Heon-Jeong Lee, Hyun Goo Woo.
Figure 1.
Manhattan plot of genome-wide association results for sleeplessness symptom of bipolar mania susceptibility loci. The chromosomal position is shown along the x-axis, the -log (p-value) for each single nucleotide polymorphism is shown along the y-axis. Horizontal line indicates the p<10-4 threshold.
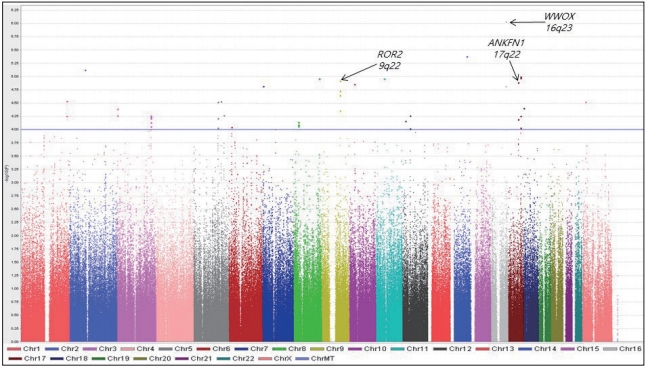
Figure 2.
Haplotype association plots for the eight ROIs in the case-control analysis of GWA. Proxy SNPs with regional recombination rates in the eight ROIs are shown as determined by SNAP with HapMap 1000 Genomes Pilot 1 data (A-H, top). The regional LDs around the eight ROIs are shown by Haploview using HapMap CEU+TSI(R2) data (A-H, bottom). The D’ scores for each paired SNP are indicated by the color scheme. The recombination rate (pale blue line), the lowest p-value SNP (the biggest diamond mark), and ROI span (dotted vertical line) are indicated. ROI: region of interest, GWA: genome-wide association, SNP: single nucleotide polymorphism, LD: linkage disequilibrium.
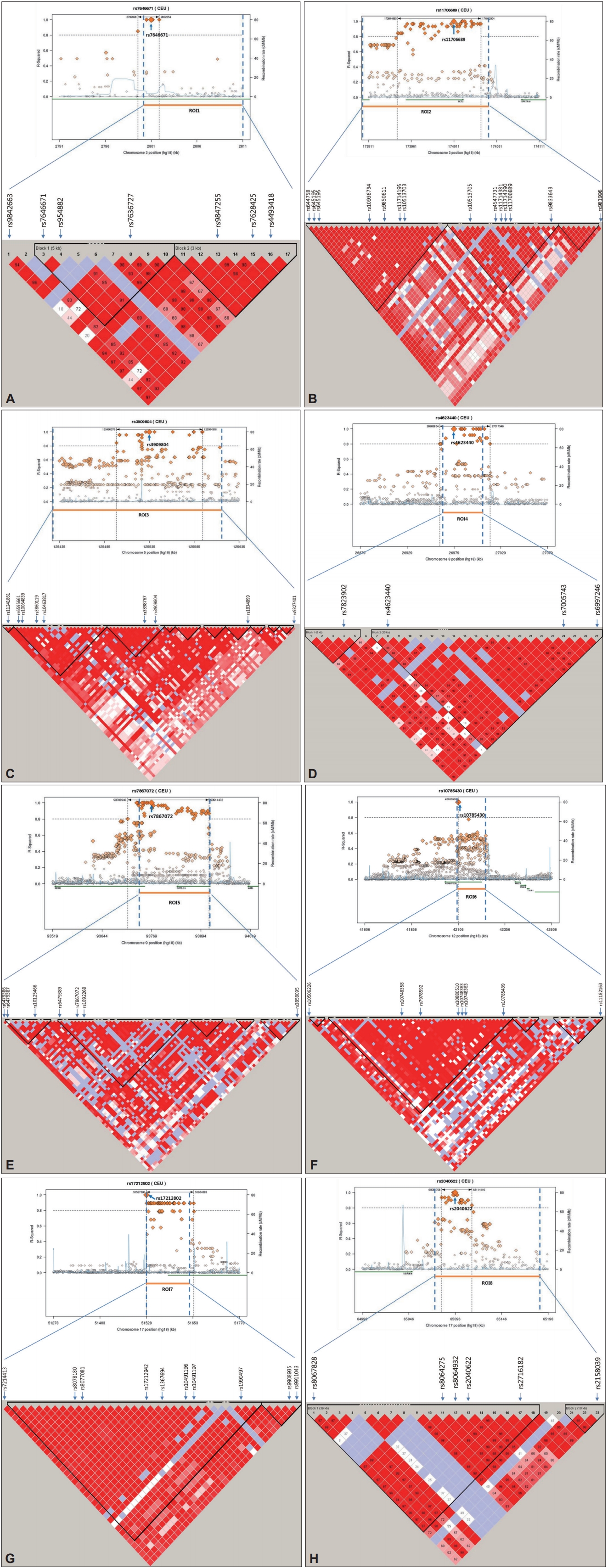
Table 1.
The enrichment findings of the functional enrichment analysis within imputed data of genome-wide association study
REFERENCES
1. Perlis RH, Ostacher MJ, Patel JK, Marangell LB, Zhang H, Wisniewski SR, et al. Predictors of recurrence in bipolar disorder: primary outcomes from the Systematic Treatment Enhancement Program for Bipolar Disorder (STEP-BD). Am J Psychiatry 2006;163:217–224.


2. Fagiolini A, Frank E, Axelson DA, Birmaher B, Cheng Y, Curet DE, et al. Enhancing outcomes in patients with bipolar disorder: results from the Bipolar Disorder Center for Pennsylvanians Study. Bipolar Disord 2009;11:382–390.



3. Akiskal HS, Bourgeois ML, Angst J, Post R, Möller H, Hirschfeld R. Reevaluating the prevalence of and diagnostic composition within the broad clinical spectrum of bipolar disorders. J Affect Disord 2000;59 Suppl 1:S5–S30.


4. Judd LL, Akiskal HS. The prevalence and disability of bipolar spectrum disorders in the US population: re-analysis of the ECA database taking into account subthreshold cases. J Affect Disord 2003;73:123–131.


5. Kessler RC, Berglund P, Demler O, Jin R, Merikangas KR, Walters EE. Lifetime prevalence and age-of-onset distributions of DSM-IV disorders in the National Comorbidity Survey Replication. Arch Gen Psychiatry 2005;62:593–602.


6. Oswald P, Souery D, Kasper S, Lecrubier Y, Montgomery S, Wyckaert S, et al. Current issues in bipolar disorder: a critical review. Eur Neuropsychopharmacol 2007;17:687–695.


7. Pini S, de Queiroz V, Pagnin D, Pezawas L, Angst J, Cassano GB, et al. Prevalence and burden of bipolar disorders in European countries. Eur Neuropsychopharmacol 2005;15:425–434.


8. Goldberg JF, Harrow M. Consistency of remission and outcome in bipolar and unipolar mood disorders: a 10-year prospective follow-up. J Affect Disord 2004;81:123–131.


9. Harvey AG. Sleep and circadian rhythms in bipolar disorder: seeking synchrony, harmony, and regulation. Am J Psychiatry 2008;165:820–829.


10. Dinges DF, Pack F, Williams K, Gillen KA, Powell JW, Ott GE, et al. Cumulative sleepiness, mood disturbance, and psychomotor vigilance performance decrements during a week of sleep restricted to 4-5 hours per night. Sleep 1997;20:267–277.

11. Drake CL, Roehrs TA, Burduvali E, Bonahoom A, Rosekind M, Roth T. Effects of rapid versus slow accumulation of eight hours of sleep loss. Psychophysiology 2001;38:979–987.


12. Harvey AG. A transdiagnostic approach to treating sleep disturbance in psychiatric disorders. Cogn Behav Ther 2009;38 Suppl 1:35–42.


13. Wehr TA, Sack DA, Rosenthal NE. Sleep reduction as a final common pathway in the genesis of mania. Am J Psychiatry 1987;144:201–204.


14. Kripke DF, Nievergelt CM, Joo E, Shekhtman T, Kelsoe JR. Circadian polymorphisms associated with affective disorders. J Circadian Rhythms 2009;7:2.



15. Mansour HA, Wood J, Chowdari KV, Dayal M, Thase ME, Kupfer DJ, et al. Circadian phase variation in bipolar I disorder. Chronobiol Int 2005;22:571–584.


16. McClung CA. Circadian genes, rhythms and the biology of mood disorders. Pharmacol Ther 2007;114:222–232.



17. McCarthy MJ, Nievergelt CM, Kelsoe JR, Welsh DK. A survey of genomic studies supports association of circadian clock genes with bipolar disorder spectrum illnesses and lithium response. PLoS One 2012;7:e32091.



18. Schreiner R, Mirisch S, Vesely Z, Wiegand MH. Sleep and sleep-wake cycle in an 81-year-old patient with de novo ultra-rapid cycling bipolar disorder. Eur Arch Psychiatry Clin Neurosci 2001;251:29–31.


19. Wehr TA, Sack D, Rosenthal N, Duncan W, Gillin JC. Circadian rhythm disturbances in manic-depressive illness. Fed Proc 1983;42:2809–2814.

20. Harvey AG, Mullin BC, Hinshaw SP. Sleep and circadian rhythms in children and adolescents with bipolar disorder. Dev Psychopathol 2006;18:1147–1168.


21. Lee HJ, Son GH, Geum D. Circadian rhythm hypotheses of mixed features, antidepressant treatment resistance, and manic switching in bipolar disorder. Psychiatry Investig 2013;10:225–232.



22. Cho CH, Lee HJ. Why do mania and suicide occur most often in the Spring? Psychiatry Investig 2018;15:232–234.



23. Jee HJ, Cho CH, Lee YJ, Choi N, An H, Lee HJ. Solar radiation increases suicide rate after adjusting for other climate factors in South Korea. Acta Psychiatr Scand 2017;135:219–227.


24. Moon JH, Cho CH, Son GH, Geum D, Chung S, Kim H, et al. Advanced circadian phase in mania and delayed circadian phase in mixed mania and depression returned to normal after treatment of bipolar disorder. EBioMedicine 2016;11:285–295.



25. Cho CH, Moon JH, Yoon HK, Kang SG, Geum D, Son GH, et al. Molecular circadian rhythm shift due to bright light exposure before bedtime is related to subthreshold bipolarity. Sci Rep 2016;6:31846.



26. American Psychiatric Association. Diagnostic and statistical manual of mental disorders (DSM-5®). Washington DC: American Psychiatric Publishing, 2013.
27. Sklar P, Smoller JW, Fan J, Ferreira MA, Perlis RH, Chambert K, et al. Wholegenome association study of bipolar disorder. Mol Psychiatry 2008;13:558–569.



28. Smith EN, Koller DL, Panganiban C, Szelinger S, Zhang P, Badner JA, et al. Genome-wide association of bipolar disorder suggests an enrichment of replicable associations in regions near genes. PLoS Genet 2011;7:e1002134.



29. Cho CH, Kim S, Geum D, Lee HJ. Association analysis of ANK3 variants with bipolar disorder in the Korean population. Nord J Psychiatry 2017;71:245–249.


30. Kim S, Cho CH, Geum D, Lee HJ. Association of CACNA1C variants with bipolar disorder in the Korean population. Psychiatry Investig 2016;13:453–457.



31. Lett TA, Zai CC, Tiwari AK, Shaikh SA, Likhodi O, Kennedy JL, et al. ANK3, CACNA1C and ZNF804A gene variants in bipolar disorders and psychosis subphenotype. World J Biol Psychiatry 2011;12:392–397.


32. Lee HJ, Woo HG, Greenwood TA, Kripke DF, Kelsoe JR. A genome-wide association study of seasonal pattern mania identifies NF1A as a possible susceptibility gene for bipolar disorder. J Affect Disord 2013;145:200–207.


33. Cho CH, Lee HJ, Woo HG, Choi JH, Greenwood TA, Kelsoe JR. CDH13 and HCRTR2 may be associated with hypersomnia symptom of bipolar depression: a genome-wide functional enrichment pathway analysis. Psychiatry Investig 2015;12:402–407.



34. Nurnberger JI Jr, Blehar MC, Kaufmann CA, York-Cooler C, Simpson SG, Harkavy-Friedman J, et al. Diagnostic interview for genetic studies. Rationale, unique features, and training. NIMH Genetics Initiative. Arch Gen Psychiatry 1994;51:849–859.discussion 863-864.


35. Sanders AR, Levinson DF, Duan J, Dennis JM, Li R, Kendler KS, et al. The Internet-based MGS2 control sample: self report of mental illness. Am J Psychiatry 2010;167:854–865.



36. Potash JB, Toolan J, Steele J, Miller EB, Pearl J, Zandi PP, et al. The bipolar disorder phenome database: a resource for genetic studies. Am J Psychiatry 2007;164:1229–1237.


37. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007;81:559–575.



38. Bush WS, Moore JH. Chapter 11: Genome-wide association studies. PLoS Comput Biol 2012;8:e1002822.



39. Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005;21:263–265.


40. Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet 2009;5:e1000529.



41. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44–57.


42. Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 2007;447:661–678.



43. Baum AE, Hamshere M, Green E, Cichon S, Rietschel M, Noethen MM, et al. Meta-analysis of two genome-wide association studies of bipolar disorder reveals important points of agreement. Mol Psychiatry 2008;13:466–467.



44. Psychiatric GWAS Consortium Bipolar Disorder Working Group. Largescale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet 2011;43:977–983.



45. Paige AJ, Taylor KJ, Taylor C, Hillier SG, Farrington S, Scott D, et al. WWOX: a candidate tumor suppressor gene involved in multiple tumor types. Proc Natl Acad Sci U S A 2001;98:11417–11422.



46. Sze CI, Su M, Pugazhenthi S, Jambal P, Hsu LJ, Heath J, et al. Down-regulation of WW domain-containing oxidoreductase induces Tau phosphorylation in vitro. A potential role in Alzheimer’s disease. J Biol Chem 2004;279:30498–30506.


47. Chen ST, Chuang JI, Wang JP, Tsai MS, Li H, Chang NS. Expression of WW domain-containing oxidoreductase WOX1 in the developing murine nervous system. Neuroscience 2004;124:831–839.


48. Mallaret M, Synofzik M, Lee J, Sagum CA, Mahajnah M, Sharkia R, et al. The tumour suppressor gene WWOX is mutated in autosomal recessive cerebellar ataxia with epilepsy and mental retardation. Brain 2014;137:411–419.


49. Wang HY, Juo LI, Lin YT, Hsiao M, Lin JT, Tsai CH, et al. WW domaincontaining oxidoreductase promotes neuronal differentiation via negative regulation of glycogen synthase kinase 3β. Cell Death Differ 2012;19:1049–1059.


50. Doble BW, Woodgett JR. GSK-3: tricks of the trade for a multi-tasking kinase. J Cell Sci 2003;116:1175–1186.



51. Abe M, Herzog ED, Block GD. Lithium lengthens the circadian period of individual suprachiasmatic nucleus neurons. Neuroreport 2000;11:3261–3264.


52. Baştürk M, Karaaslan F, Esel E, Sofuoğlu S, Tutuş A, Yabanoğlu I. Effects of short and long-term lithium treatment on serum prolactin levels in patients with bipolar affective disorder. Prog Neuropsychopharmacol Biol Psychiatry 2001;25:315–322.


53. Campbell SS, Gillin JC, Kripke DF, Janowsky DS, Risch SC. Lithium delays circadian phase of temperature and REM sleep in a bipolar depressive: a case report. Psychiatry Res 1989;27:23–29.


54. Seggie J, Werstiuk ES, Grota L. Lithium and circadian patterns of melatonin in the retina, hypothalamus, pineal and serum. Prog Neuropsychopharmacol Biol Psychiatry 1987;11:325–334.


55. Klein PS, Melton DA. A molecular mechanism for the effect of lithium on development. Proc Natl Acad Sci U S A 1996;93:8455–8459.



56. Stambolic V, Ruel L, Woodgett JR. Lithium inhibits glycogen synthase kinase-3 activity and mimics wingless signalling in intact cells. Curr Biol 1996;6:1664–1668.


57. Li X, Bijur GN, Jope RS. Glycogen synthase kinase-3beta, mood stabilizers, and neuroprotection. Bipolar Disord 2002;4:137–144.



58. Paganoni S, Bernstein J, Ferreira A. Ror1-Ror2 complexes modulate synapse formation in hippocampal neurons. Neuroscience 2010;165:1261–1274.


59. Ban HJ, Kim SC, Seo J, Kang HB, Choi JK. Genetic and metabolic characterization of insomnia. PLoS One 2011;6:e18455.



60. Smith EN, Bloss CS, Badner JA, Barrett T, Belmonte PL, Berrettini W, et al. Genome-wide association study of bipolar disorder in European American and African American individuals. Mol Psychiatry 2009;14:755–763.



- TOOLS







